Project Milestone Report
Project Milestone
Andrew Kubaney (ajkubane)
Haoyu Zhang (haoyuzha)
New URL:
https://15418-fall22-team-kz.github.io/project-campfire-website/
Progress on Original Schedule
- Week 1 (11/7 to 11/13): For the rest of this week, we will focus on finalizing the algorithm for the chemical simulation (including the simplifying assumptions necessary). Then, we will begin converting the particle simulation to the chemical particle simulation (switch to electrostatic force, introduce collisions).
- COMPLETED: Switched to electrostatic force.
- Week 2 (11/14 to 11/20): We will finish converting the particle simulation (add reactions), beginning work on the concentration simulation (first looking at simpler algorithms than the Gillespie algorithm).
- COMPLETED: We implemented a simple concentration simulation (we determined that the more complex concentration simulations were not necessary).
- IN PROGRESS: We are still finalizing our implementation of collisions and reactions. We have most of the logic implemented, and the last thing to do is to implement a few physics concepts for collisions and decomposition reactions.
- Week 3 (11/21 to 11/27): We will finish work on the concentration simulation (if the simpler algorithms are very easy to implement, then we may try to implement the Gillespie algorithm, and begin working on the sequential multi-granularity simulation (combining the two granularities into one scene).
- COMPLETED: We began work on the combined granularity simulation.
- Week 4 (11/28 to 12/4): Finish working on the sequential multi-granularity simulation (Note, this week is a little bit light on work because Andrew has many graduate school applications due this week. We will make up for it by working extra the next week).
- IN PROGRESS: We are close to finishing the combined granularity simulation. The last thing to do is to finish implementing collisions and decomposition reactions. As mentioned above, these tasks are mostly finished.
- Week 5 (12/5 to 12/11): Parallelize the simulation with OpenMP. Find a simple chemical reaction with known parameters (experimentally determined in the lab), and use this reaction to test the correctness of our simulation.
- Week 6 (12/12 to 12/18): Perform analysis of the parallel implementation (on the GHC machines). If time allows, perform more complex analysis, look at more complex chemical scenes, and/or measure performance on the PSC machines. Finally, collect the results into the final documentation and prepare for the final presentation.
- 12/17/2022: Submit the final report
- 12/18/2022: Project poster session
Revised Schedule
We break the remaining time down into half-week increments. There are five half-week increments remaining before the project deadline:
- Half Week 1:
- Finish sequential version (Andrew)
- Understand sequential version (Haoyu)
- Half Week 2:
- Test sequential version on sample scenarios (Haoyu)
- Naive parallelization with OpenMP (Andrew)
- Half Week 3:
- Improve OpenMP parallelization, particularly in collision handling and generation of particles from concentration regions (Andrew and Haoyu)
- Half Week 4:
- Analyze code performance on GHC machines (Andrew)
- Analyze code performance on PSC machines (Haoyu)
- Half Week 5:
- Write report (Andrew)
- Create poster (Haoyu)
Work Completed So Far
1. Combined Granularity Simulation
We have implemented most of the combined concentration-particle simulation. There are several differences between the algorithm that we settled on and the algorithm we described in the project proposal. For each iteration, our current algorithm does the following (note, all parts are implemented except those denoted as NOT FINISHED):
- For each concentration region, on the outer edges of the region, use the concentrations to generate particles. These particles are then added to the rest of the particles. In this way, the concentration regions are able to affect the particle regions.
- Simulate the electrostatic forces and update the particles.
- For each particle, determine if there are any other colliding particles.
- If so, if a forward reaction is possible, with a certain probability, allow a forward reaction to occur.
- Otherwise, update the colliding particles (NOT FINISHED).
- For each particle, if a backward reaction is possible, allow a backward reaction to occur (NOT FINISHED).
- For each particle, if it is in a concentration region, fold the particle into the concentration for that region.
- For each concentration region, update the concentrations based on the forward and backward reaction.
Note, we also made some simplifications. First, instead of allowing for arbitrary amounts and types of reactions, we opted to focus on one type of forward and one type of backward reaction. We made this decision because we realized that the work required to generalize the simulation would not significantly augment the final result of our project. Another simplification that we made was to abstract away the concepts of temperature and activation energy. Ultimately, we believe that these concepts overcomplicated our algorithm and that we will be able to replicate the behavior of the more complicated algorithm with carefully chosen constants.
2. Random chemical scenes generation and data visualization
As we were provided with different test scenes in assignment 3 and 4, we also need some pre-defined scenes for the initial states of our chemical simulation, and to explore potential parallelization methods we may apply for different input patterns. Specifically, the “patterns” we’re referring to here are distinct distributions of concentration regions as well as the concentration of the three types of molecules within the regions.
We use Python to generate scenes with concentration regions in fixed locations and molecule with random locations. And also we use Python to visualize the data generated by each simulation step. A sample visualization of an initial step looks like this:
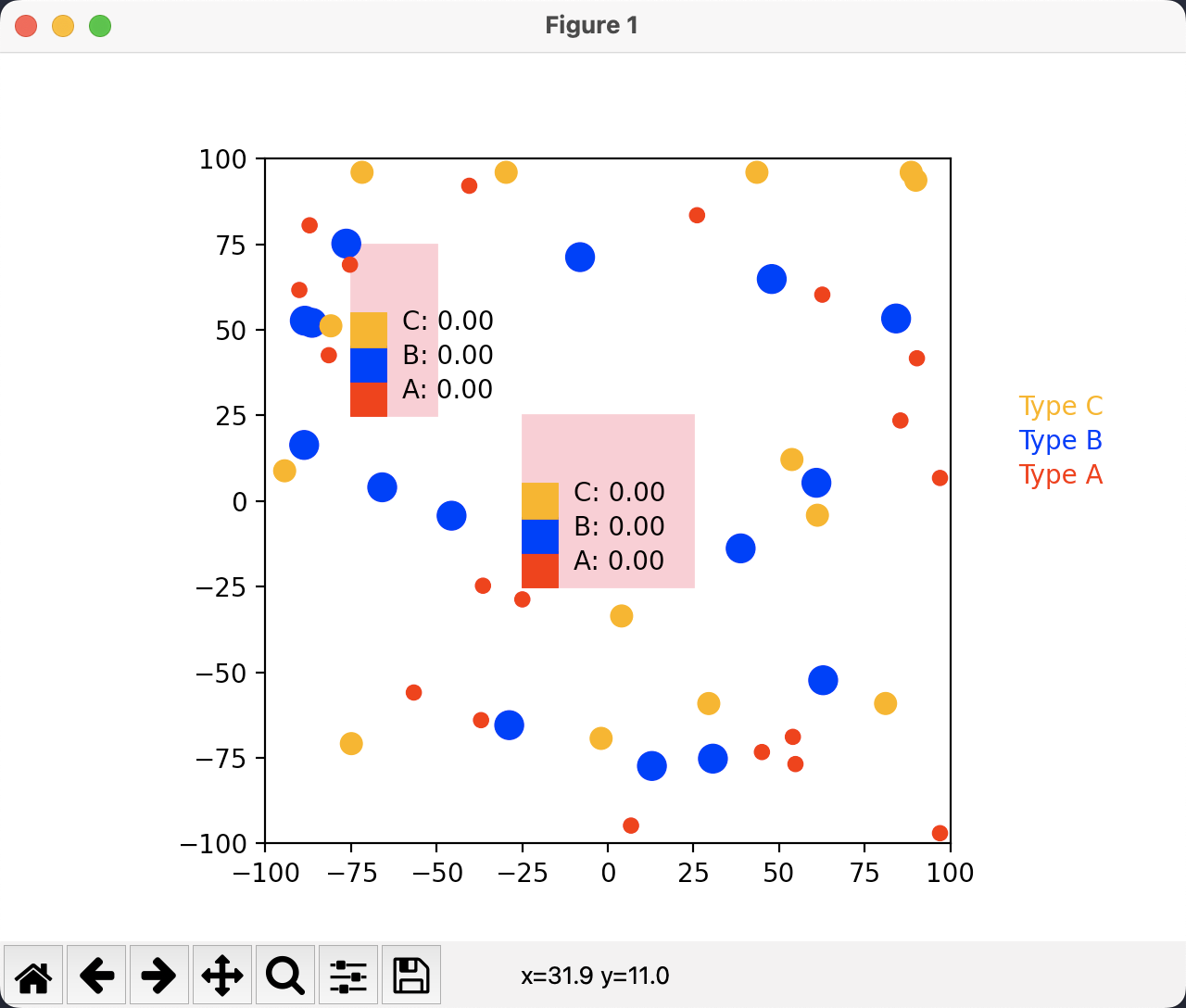
The size of this sample scene is 200 by 200. We can observe that we have three types of molecules (as discussed above) which are represented as circles with different color identifying their type and different radius as defined. The pink rectangle are the concentration areas. Individual molecules will not show up in theses regions. Instead, they are treated as a whole indicated by the concentrations.
For illustration, each pink region has three legends specifying concentration of the different types of molecules within that region. The numbers are 0.00 because they are rounded off. Actual simulation scenes are expected to have higher concentrations. And note that these legends are not used in our final visualization. They are used just for illustration and test purposes.
Now our script is capable of reading and parsing the intermediate simulation results and draw the plot like the one shown above. We plan to link up the images and create GIFs to showcase the simulations done by our program. More visualization details and effects are expected to be done in the future.
Progress on Original Goals/Deliverables
Our goals from the proposal are displayed below. For the most part, we believe that we will be able to provide all of our deliverables. There are a few deliverables that we decided to remove, as we believe that they will not significantly augment our final project (relative to the amount of work that they would necessitate).
Plan to Achieve
- Convert the particle simulation to the chemical molecule simulation.
- CLOSE TO COMPLETION: As described above, the only things left to implement are some physics concepts for collisions and decomposition reactions.
- Implement simulation for concentration regions.
- COMPLETED: As described above, we have fully implemented the concentration simulations.
- Allow for scenes to have both molecule and concentration regions.
- CLOSE TO COMPLETION: As described above, the only things left to implement are some physics concepts for collisions and decomposition reactions.
- Parallelize this code with OpenMP.
- INCOMPLETE BUT FEASIBLE: A simple parallelization will be straightforward. More effective parallelization will require us to more closely examine our collision logic.
- Use a simple chemical reaction (one idea is bleach and red dye) to test the correctness of our implementation.
- CHANGED: It will be difficult to match our simulation exactly to experimental data. Instead, we will focus on more high-level correctness of our code, ensuring that the high-level behaviors (such as diffusion of particles from high to low concentration regions, reactions, and collisions) are occurring as expected.
- Analyze the code. Measure the performance (speedup, memory accesses, etc.) on the GHC machines.
- INCOMPLETE BUT FEASIBLE: Once we have the parallelized version, this will be relatively straightforward.
Hope to Achieve
- More advanced profiling of threads (looking at data, sync, busy times).
- INCOMPLETE BUT FEASIBLE: Once we have the parallelized version, this will be relatively straightforward.
- Make more advanced scenes with more chemical reactions.
- REMOVED: As described above, we decided to simplify our simulation to involve a single forward reaction and a single backwards reaction. To reiterate, this was done because we believe that the complexity of generalizing the reactions is not worth the benefit (within the scope of our project). As such, this deliverable no longer makes sense.
- Test our code on the PSC machines. Compare the performance to the performance on the GHC machines.
- INCOMPLETE BUT FEASIBLE: Once we have the parallelized version, this will be relatively straightforward.
Revised Goals/Deliverables
The following represent a revised version of our remaining goals and deliverables.
- Finish combined granularity simulation.
- Test for high-level correctness.
- Complete a naive parallelization with OpenMP.
- Improve the parallelization, specifically as it pertains to particle generation from concentration regions and collisions.
- Analyze the code on GHC machines (including per-thread profiles).
- Analyze the code on PSC machines (including per-thread profiles).
- Write report and generate GIF/figures for demo.
Poster Session
Our plan for the poster session remains unchanged from our proposal. We plan to show a visual demonstration of our multi-granularity chemical simulation in the form of a GIF. We will also display speedup graphs with our code on multiple processors vs. a single processor.
Remaining Unknowns
There are three remaining unknowns for our project.
- We are still deciding the best way to demonstrate the correctness of our simulation. As described above, for the scope of this project, we have determined that it is not feasible to mimic experimental data. Our current plan is to choose specific scenes where we can predict the high-level outcome and compare our expectations to the results from the code.
- We anticipate that the parallelization of the particle generation from the concentration regions will not be easy. While randomly generating particles in parallel is not difficult, we want to find a way to generate these particles in a way that does not depend on execution order. Our sequential code utilizes a seeded random number generator, which is not straightforward to generalize to multiple processors.
- The parallelization of our current collision resolution code is not straightforward. If the collision of two particles results in a reaction, we no longer want the reactant particles to be involved in collisions. This creates inherent dependencies which are difficult to resolve when multiple processors are involved.